NSF Bioinformatics Grant awarded to CIS/Biology

NSF's Division of Environmental Biology has awarded Dr. John Conery and Dr. Joe Thornton (Biology) $353,000 to study Mixed-Model Phylogenetic Methods for Evolutionarily Heterogeneous Data.
According to PhD student and fellow researcher Bryan Kolaczkowski, "Heterogeneity in the evolutionary process is a hot topic right now, as current models assume a large degree of homogeneity, yet we know that this assumption is wrong. In addition, our earlier work - and the work of others - has shown that failure to incorporate hetergeneous evolutionary features can bias current methods toward the wrong inference." To counter this bias, the team has been developing a heterogeneous mixed-model approach to describe the evolutionary process. The idea of a mixed-model is to describe the data using more than one parameterization. This allows the software to incorporate the fact that the sample sites may experience slightly different evolutionary models.
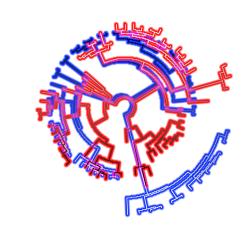
Modeling Heterotachy: different data subsets are allowed to evolve at different evolutionary rates, which can change as lineages evolve.
The grant will fund Kolaczkowski's participation through the end of his Phd program and subsequently as a post-doc. The grant will also purchase a new computational cluster to provide the intensive computing resources required for this type of modeling. The cluster will consist of eight nodes of dual-processor X-Serve from Apple, running OS X Server 10.4, and the cluster management system called INquiry, which comes bundled with a wide range of bioinformatics software. The team plans to incorporate their new model into the INquiry framework to make it readily available to other bioinformatics researchers.
The complete text of the latest article in support of the grant is available in Nature, vol 431, no 7011, pp. 980-984. More information can be obtained at the websites of Dr. Joe Thornton and Dr. John Conery